Playing Around with RNA
Instead of commenting on other's people's work ... (for that go to some other great science blogs) ... my new blog will try to have more original stuff.
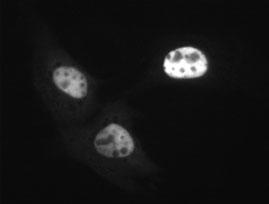
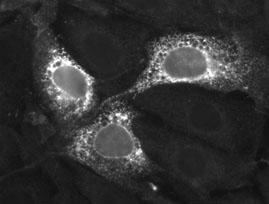
First up, the synthetic gene. As you can tell by my URL and the title of my blog, my interest currently lies with mRNA. To figure out how mRNA gets localized, I've synthesized some in a test tube and microinjected it into nuclei of tissue culture cells (click here for some idea on what a tissue culture cell looks like). Above are two pics of the same field cells. Displayed in the second picture is what is called an injection marker, or a fluorescent substance to mark which cells are injected. Since this marker (FITC-70kD dextran) is too big to cross the nuclear membrane, it demonstrates that in all three cells, all the injection fluid entered the nucleus and not the rest of the cell (aka the cytoplasm).
But studying mRNA is a pain in the butt. RNA gets degraded quite easily. Also the way cells handle RNA remains unclear. One thing that I did to check that the injected cells are handling the RNA correctly, was to alter the mRNA so that it should have all the right signals to be translated into protein. To do this the mRNA must start with a Cap structure, then a leader sequence, followed by a Kozak sequence (named after Marilyn Kozak), a stop sequence and a polyA-tail (see here and here for more on mRNA). In addition the mRNA was modified so that it produces protein with a "FLAG tag" sequence that can be detected by fluorescent antibodies (see top picture) and a signal sequence, that targets the encoded protein to the endoplasmic reticulum (ER). Now if I try to probe the same cells for translated protein (encoded by my mRNA) voila, I see that the FLAG tag in a reticular type pattern reminiscent of the ER (top picture) in the same cells that I injected (see bottom pic).


<< Home